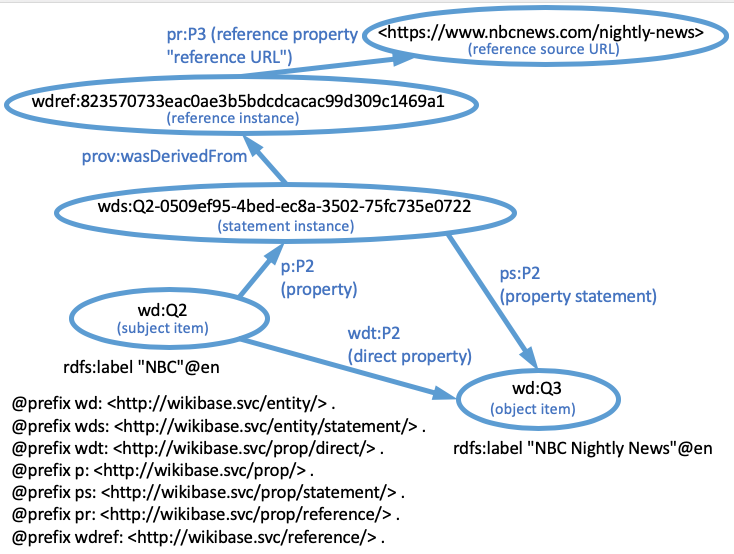
This post is very focused on the details of two
Biodiversity Information Standards (TDWG) standards as they relate to Linked Data and graph models. If you are generally interested in approaches to Linked Data graph modeling, you might find it interesting. Otherwise, if you aren't into TDWG standards, you may zone out.
Background
The TDWG Darwin Core and Access to Biological Collection Data (ABCD) standards
Access to Biological Collection Data (ABCD) is a standard of Biodiversity Information Standards (TDWG). It is classified as a "Current Standard" but is in a special category called "2005" standard because it was ratified just before the present
TDWG by-laws (which specify the details of the standards development process) were adopted in 2006. Originally, ABCD was defined as an XML schema that could be used to validate XML records that describe biodiversity resources. The various versions of the ABCD XML schema can be found in the
ABCD GitHub repository.
Darwin Core (DwC) is a current standard of TDWG that was ratified in 2009. It is modeled after
Dublin Core, with which it shares many similarities. Biodiversity data can be transmitted in several ways: as
simple spreadsheets, as
XML, and as
text files structured in a form known as a Darwin Core Archive.
Nearly all of the more than 1.3 billion records in the
Global Biodiversity Information Facility (GBIF) have been marked up in either DwC or ABCD.
My role in Darwin Core
For some time I've been interested in the possibility of using Darwin Core terms as a way to transmit biodiversity data as Linked Open Data (LOD). That interest has manifested itself in my being involved in three ways with the development of Darwin Core:
All three of these official changes to Darwin Core were approved by decision of the TDWG Executive Committee on October 26, 2014. Along with Cam Webb, I also was involved in an unofficial effort called
Darwin-SW (DSW) to develop an RDF ontology to create the graph model and object properties that were missing from the Darwin Core vocabulary. (For details, see
http://dx.doi.org/10.3233/SW-150203; open access at
http://bit.ly/2dG85b5.) More on that later...
I've had no role with ABCD and honestly, I was pretty daunted about the prospect of plowing through the XML schema to try to understand how it worked. However, I've recently been using some new tools Linked Data tools to explore ABCD and they have been instrumental for putting the material together for this blog. More about them later...
A common model for ABCD and Darwin Core?
Recently, a call went out to people interested in developing a common model for TDWG that would encompass both ABCD and DwC. Because of my past interest in using Darwin Core terms as RDF, I joined the group, which has met online once so far. Because of my basic ignorance about ABCD, I've recently put in some time to try to understand the existing model for ABCD and how it is similar or different from Darwin Core. In the following sections, I'll discuss some issues with modeling Darwin Core, then report on what I've learned about ABCD and how it compares to Darwin Core.
Darwin Core's missing graph model
One of the things that surprises some people is that although a DwC RDF Guide exists, it is not currently possible to express biodiversity data as RDF using only terms currently in the standard.
What the RDF Guide does is to clear up how the existing terms of Darwin Core should be used and to mint some new terms that can be used for creating links between resources (i.e. to non-literal objects of triples). For example, as adopted, Darwin Core had the term
dwc:recordedBy (
http://rs.tdwg.org/dwc/terms/recordedBy) to indicate the person who recorded the occurrence of an organism. However, it was not clear whether the value of this term (i.e. the object of a triple of which the predicate was
dwc:recordedBy) should be a literal (i.e. a name string) or an IRI (i.e. an identifier denoting an agent). The RDF Guide establishes that
dwc:recordedBy should be used with a literal value, and that a new term,
dwciri:recordedBy (
http://rs.tdwg.org/dwc/iri/recordedBy) should be used to link to an IRI denoting an agent (i.e. a non-literal value). For each term in Darwin Core where it seemed appropriate for an existing term to have a non-literal (IRI) value, a
dwciri: namespace analog of that term was created. The terms affected by this decision are detailed in the
Term reference section of the guide.
So with the RDF Guide, it is now possible to express a lot of Darwin Core metadata as RDF. But at the time of the adoption of the RDF Guide there were no existing DwC terms that linked instances of the DwC classes (i.e. object properties), so there was no way to fully express a dataset as RDF. (Another way of saying this is that Darwin Core did not have a graph model for its classes.) It seems like there should be a simple solution to that problem: just define some object properties to connect the classes. But as Joel Sachs and I describe in
a recent book chapter, that's not as simple as it seems. In section 3.2 of the chapter, we show how users with varying interests may want to use graph models that are more or less complex, and that inconsistencies on those models makes it difficult to query across datasets that use different models.
The Darwin Core RDF Guide was developed not long after a bruising, year-long online discussion about modeling Darwin Core (see
this page for a summary of the gory details). It was clear that if we had planned to include a graph model and the necessary object properties, the RDF Guide would probably never get finished. So it was decided to create the RDF Guide to deal with the existing terms and leave the development of a graph model as a later effort.
Darwin-SW's graph model
After the exhausting online discussion (argument?) about modeling Darwin Core, I was so burned out on the subject, I had decided that I was basically done with that subject. However, Cam Webb, the eternal optimist, contacted me and said that we should just jump in and try to create a QL-type ontology that had the missing object properties. (See "For further reference" at the end for definitions of "ontology").
What made that project feasible was that despite the rancor of the online discussion, there actually did seem to be some degree of consensus about a model based on historical work done 20 years earlier. Rich Pyle had laid out a diagram of a model that we were discussing and Greg Whitbread noted that it was quite similar to the Association of Systematics Collections (ASC) model of 1993. All Cam and I really had to do was to create object properties to connect all of the nodes on Rich's diagram. We worked on it for a couple of weeks and the first draft of Darwin-SW (DSW) was done!
The diagram above shows the DSW graph model overlaid upon the ACS entity-relation (ER) diagram. I realize that it's impossible to see the details in this image, but you can download a poster-sized PowerPoint diagram from
this page to see the details.
DSW differs a little from the ASC model in that it includes two Darwin Core classes (dwc:Organism and dwc:Occurrence) that weren't dealt with in the ACS model. Since the ACS model dealt only with museum specimens, it did not include the classes of Darwin Core that were developed later to deal with repeated records of the same organism, or records documented by forms of evidence other than specimens (i.e. human and machine observations, media, living specimens, etc.). But other than that, the DSW model is just a simplified version of the ACS model.
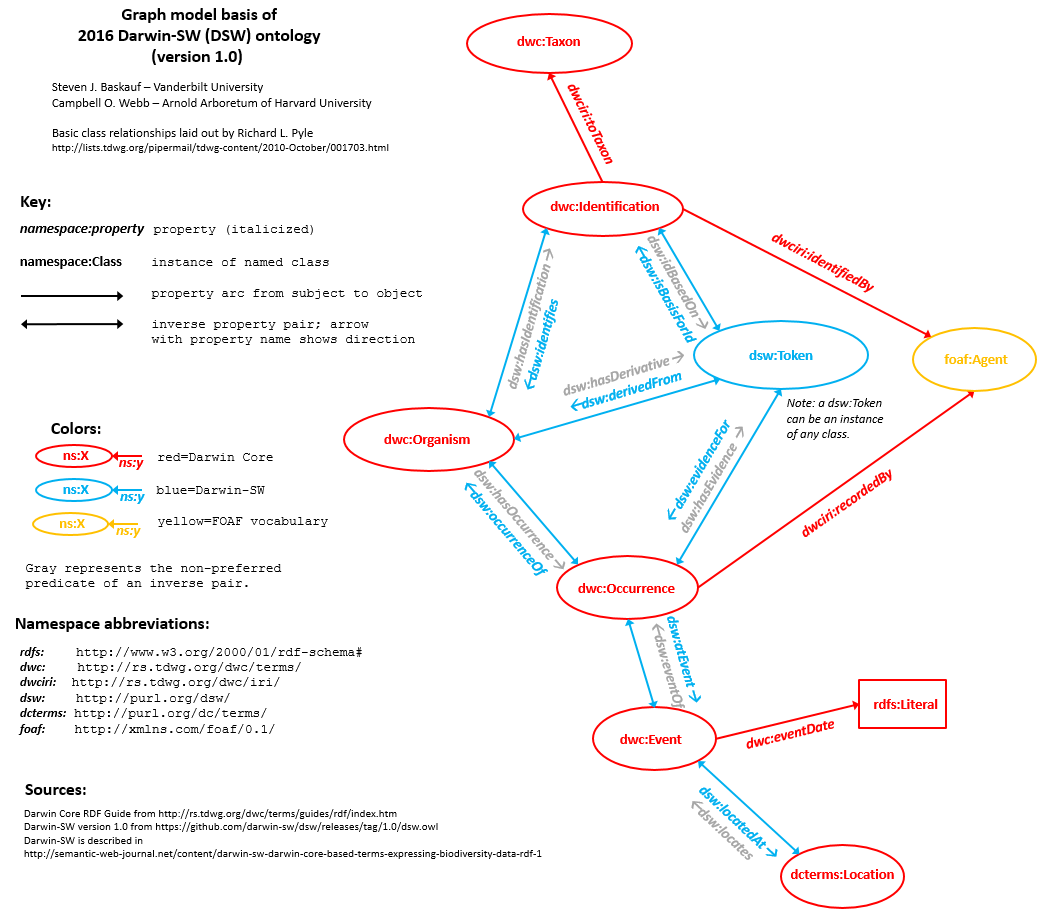
The diagram above shows the core of the DSW graph model (available
poster-sized here if you have trouble seeing the details). The six red bubbles are the six major classes defined by Darwin Core. The yellow bubble is FOAF's Agent class, which can be linked DwC classes by two terms from the
dwciri: namespace. The object of
dwc:eventDate is a literal, and
dwciri:toTaxon links to some yet-to-be-fully-described taxon-like entity that will hopefully be fleshed out by a successor to the
Taxon Concept Transfer Schema (TCS) standard, but whose place is currently being held by the
dwc:Taxon class. The seven object properties printed in blue are DSW's attempt to fill in the object properties that are missing from the Darwin Core standard.
The blue bubble, dsw:Token, is one of the few classes that we defined in DSW instead of borrowing from elsewhere. We probably should have called it dsw:Evidence, because "evidence" is what it represents, but too late now. I will talk more about the Token class in the next section.
What's an Occurrence???
One of the longstanding and vexing questions of users of Darwin Core is "what the heck is an occurrence?" The origin of dwc:Occurrence predates my involvement with TDWG, but I believe that its creation was to solve the problem of overlap of terms that applied to both observations and preserved specimens. For example, you could have terms called dwc:observer and dwc:collector, with observer being used with observations and collector being used with specimens. Similarly, you could have dwc:observationRemarks for observations and dwc:collectionRemarks for specimens. But fundamentally, both an observer and a collector are creating a record that an organism was at some place at some time, so why have two different terms for them? Why have two separate remarks term when one would do? So the dwc:Occurrence class was created as an artificial class to organize terms that applied to both specimens and observations (like the two terms dwc:recordedBy and dwc:occurrenceRemark that replace the four terms above). Any terms that applied to only specimens (like dwc:preparations and dwc:disposition) were thrown in the Occurrence group as well.
So for some time, dwc:Occurrence was considered by many to be a sort of superclass for both specimens and observations. However, its definition was pretty murky and a bit circular. Prior to our clarification of class definitions in October 2014, the definition was "The category of information pertaining to evidence of an occurrence in nature, in a collection, or in a dataset (specimen, observation, etc.)." After the class definition cleanup, it was "An existence of an Organism (sensu http://rs.tdwg.org/dwc/terms/Organism) at a particular place at a particular time." That's still a bit obtuse, but appropriate for an artificial class whose instances document that an organism was at a certain place at a certain time.
What DSW does is to clearly separate the artificial Occurrence class from the actual resources that serve to document that the organism occurred. The dsw:Token class is a superclass for any kind of resource that can serve as evidence for the Occurrence. The class name, Token, comes from the fact that the evidence also has a dsw:derivedFrom relationship with the organism that was documented -- it's a kind of token that represents the organism. There is no particular limit to what type of thing can be a token; it can be a preserved specimen, living specimen, image, machine record, DNA sequence, or any other kind of thing that can serve as evidence for an occurrence and is derived in some way from the documented organism. The properties of Tokens are any properties appropriate for any class of evidence: dwc:preparation for preserved specimens, ac:caption for images, etc.
Investigating ABCD
I mentioned that recently I gained access to some relatively new Linked Data tools for investigating ABCD. One that I'm really excited about is a
Wikibase instance that is loaded with the ABCD terminology data. If you've read any of my
recent blog posts, you'll know that I'm very interested in learning how Wikibase can be used as a way to manage Linked Data. So I was really excited both to see how the ABCD team had fit the ABCD model into the Wikibase model and also to be able to use the built-in
Query service to explore the ABCD model.
The other useful thing that I just recently discovered is an
ABCD OWL ontology document in RDF/XML serialization. It was loaded into the ABCD GitHub repo only a few days ago, so I'm excited to be able to use it as a point of comparison with the Wikibase data. I've loaded the ontology triples into the Vanderbilt Libraries' triplestore as the named graph
http://rs.tdwg.org/abcd/terms/ so that I can query it using
the SPARQL endpoint. In most of the comparisons that I've done, the results from the OWL document and the Wikibase data are identical. (As I noted in the "Time for a Snak" section of
my previous post, the Wikibase data model differs significantly from the standard RDFS model of class, range, domain, etc. So querying the two data sources requires some significant adjustments in the actual queries used in order to fit the model that the data are encoded in.)
One caveat is that
ABCD 3.0 is currently under development and the Wikibase installation is clearly marked as "experimental". So I'm assuming that the data both there and in the ontology are subject to change. Nevertheless, both of these data sources has given me a much better understanding of how ABCD models the biodiversity universe.
Term types
The
Main Page of the Wikibase installation gives a good explanation of the types of terms included in its dataset. In the description, they use the word "concept", but I prefer to restrict the use of the word "concept" to what I consider to be its standard use: for controlled vocabulary terms. (See the "For further reference" section for more on this.") So to translate their "types" list, I would say they describe one type of vocabulary (Controlled Vocabulary Q14) and four types of terms: Class Q32, Object Property Q33, Datatype Property Q34, and Controlled Term (i.e. concept) Q16.
For comparison purposes, the class and property terms in the
abcd_concepts.owl OWL ontology are typed as:
owl:Class,
owl:ObjectProperty, and
owl:DatatypeProperty. The controlled vocabularies are typed as
owl:Class rather than
skos:ConceptScheme, so subsequently the controlled vocabulary terms are typed as instances of the classes that correspond to their containing controlled vocabularies (e.g.
abcd:Female rdf:type abcd:Sex), rather than as
skos:Concept. It's a valid modeling choice, but isn't according to the recommendations of the TDWG Standards Documentation Specification. (More details about this later in the " The place of controlled vocabularies in the model" section.)
The query service makes it easy to discover what properties have actually been used with each type of term. Here is an example for Classes:
PREFIX bwd: <http://wiki.bgbm.org/entity/>
PREFIX bwdt: <http://wiki.bgbm.org/prop/direct/>
SELECT DISTINCT ?predicate ?label WHERE {
?concept bwdt:P8 bwd:Q219.
?concept bwdt:P9 bwd:Q32.
?concept bwdt:P25 ?name.
?concept ?predicate ?value.
OPTIONAL {
?genericProp wikibase:directClaim ?predicate.
?genericProp rdfs:label ?label.
}
MINUS {
?otherGenericProp wikibase:claim ?predicate.
}
}
ORDER BY ?predicate
This query is complicated a bit by the somewhat complex way that Wikibase handles properties and their labels (see
this for details), but you can see that it works by going to
https://wiki.bgbm.org/bdidata/query/ and pasting the query into the box.
One of the cool things that the Wikibase Query service allows you to do is copy the link from the browser URL bar and the link contains the query itself as part of the URL. This means that you can link directly to the query so that when you click on
the link, the query will load itself into the Query Service GUI box. So to avoid cluttering up this post with cut and paste queries, I'll just link the queries like this:
properties used with object properties,
datatype properties,
controlled terms, and
controlled vocabularies.
If you run each of the queries, you'll see that the properties used to describe the various term and vocabulary types are similar to the table shown at the bottom of the
Main Page.
Classes
One of the things I was interested in finding out about were the classes that were included in ABCD.
This query will create a table of all of the classes in ABCD 3.0 along with basic information about them. One thing that is very clear from running the query is that ABCD has a LOT more classes (57) than DwC (15). Fortunately, the classes are grouped into categories based on the core classes they are associated with. This was really helpful for me because it made it obvious to me that Gathering, Unit, and Identification were key classes in the model. The Identification class was basically the same as the
dwc:identification class of Darwin Core. The Gathering class, defined as "A class to describe a collection or observation event." seems to be more or less synonymous to the
dwc:Event class. The Unit class, defined as "A class to join all data referring to a unit such as specimen or observation record" is almost exactly how I described the
dwc:Occurrence class: an artificial class that's used to group properties that are common to specimens and observations.
Object properties
Another key thing that I wanted to know was how the ABCD 3.0 graph model compared with the DSW graph model. In order to do that, I needed to study the object properties and find out how they connected instances of classes.
As we can see from the table of term properties on the Main Page, object properties are required to have a defined range. They are not required to have a domain. Cam and I got a lot of flack when we assigned ranges and domains to object properties in DSW because of the way ranges and domains can generate unintended entailments. There is a common misconception that if one assigns a range to an object property that it REQUIRES that the object to be in instance of that class. Actually what it does is to entail that the object IS an instance of that class, whether that makes sense or not. We were OK with assigning ranges and domains in DSW because we didn't want people to use the DSW object properties to link class instances other than those that we specified in our mode - if people ignored our guidance, then they got unintended entailments. In ABCD the object properties all have names like "hasX", so if the object of a triple using the property isn't an instance of class "X", it's pretty silly to use that property. So here is makes some sense to assign ranges. Perhaps wisely, few of the ABCD object properties have the optional domain declaration. That allows those properties to be used with subject resources other than types that might have been originally envisioned without it entailing anything silly.
Instead of assigning domains, ABCD uses the property
abcd:associatedWithClass to indicate the class or classes whose instances you'd expect to have that property.
Here's a query that lists all of the object properties, their ranges, and the subject class with which they are associated. The query shows that there are a much larger number of link types (135) than DSW has. That's to be expected since there are a lot more classes. The actual number of ABCD object properties (88) is less than the number of link types because some of the object properties are used to link more than one combination of class instances.
Comparison of the DSW and ABCD graph model
 |
| Color coding described in text |
I went through the rather labor-intensive process of creating a PowerPoint diagram (above) that overlays part of the ABCD graph model on top of the DSW graph diagram that I showed previously. (There are other ABCD classes that I did't include because the diagram was too crowded and I was getting tired.) Although ABCD has a whole bunch of extra classes that don't correspond to DwC classes, the main DwC classes are have ABCD analogs that are connected in a very similar manner to the way they are connected in DSW. The resemblance is actually rather striking.
Here are a few notes about the diagram. First of all, it isn't surprising that ABCD doesn't have an Organism class that corresponds to dwc:Organism. As its name indicates, "Access to Biological Collections Data" is focused primarily on data from collections. As I learned from the fight to get dwc:Organism added to Darwin Core, collections people don't care much about repeated observations. They generally only sample an organism once since they usually kill it in the process. So they rarely have to deal with multiple occurrences linked to the same organism. However, people who track live whales or band birds care about the dwc:Organism class a lot since its primary purpose is to enable one-to-many relationships between organisms and occurrences (as opposed to having the purpose of creating some kind of semantic model of organisms).
Another obvious difference is the absence of any Location class that's separate from abcd:Gathering. Another common theme in discussing a model for Darwin Core was whether there was any need to have a dwc:Event class in addition to the dcterms:Location class, or if we could just denormalize it out of existence. In that case, the disagreement was between collections people (who often only collect at a particular location once) and people who conducted long-term monitoring of sites (who therefore had many sampling Events at one Location).
The general theme here is that people who don't have one-to-many (or many-to-many) relationships between classes don't see the need for the extra classes and omit them from their graph model. But the more diverse the kinds of datasets we want to handle with the model, the more complicated the core graph model needs to be.
The other thing that surprised me a little in the ABCD graph model was that the "Unit" was connected to the "Gathering Agent" through an instance of
abcd:FieldNumber, instead of being connected directly as does
dwciri:recordedBy. I guess that makes sense if there's a one-to-many relationship between the Unit and the FieldNumber (several Gathering Agents assign their own FieldNumber to the Unit). There are some parallels with
dwciri:fieldNumber, although it is defined to have a subject that is field notes and an object that is a
dwc:Event. (see table 3.7 in the
DwC RDF Guide). Clearly there would be some work required to harmonize DwC and ABCD in this area.
The other part of the two graph models I want to draw attention to is the area of dsw:Token.
There are two different ways of imagining the dsw:Token class. One way is to say that dsw:Token is a class that includes every kind of evidence. In that view, we enumerate the token classes we can think of, then define them using the properties associated with those kinds of evidence. The other way to think about it is to say that all of the properties that we can't join together under the banner of dwc:Occurrence get grouped under an appropriate kind of token. In that view, our job is to sort properties, and we then name the token classes as a way to group the sorted properties. These are really just two different ways of describing the same thing.
The ABCD analog of the dsw:Token class is the class abcd:TypeSpecificInformation. Its definition is: "A super class to create and link to type specific information about a unit." Recall that the definition of a Unit is "A class to join all data referring to a unit such as specimen or observation record". These definitions correspond to the "sorting out of properties" view I described above. Properties common to all kinds of evidence are organized together under the Unit class, but properties that are not common get sorted out into the appropriate specific subclass of abcd:TypeSpecificInformation.
 |
| ABCD class hierarchy |
The diagram above shows the "enumeration of types of evidence" view. In the diagram, you can see most of the imaginable kinds of specific evidence types listed as subclasses of abcd:TypeSpecificInformation. These subclasses correspond with some of the possible DwC classes that could serve as Tokens: abcd:HerbariumUnit corresponds to dwc:PreservedSpecimen, abcd:BotanicalGardenUnit corresponds to dwc:LivingSpecimen, abcd:ObservationUnit corresponds to dwc:HumanObservation, etc.
 |
| Object properties linking abcd:Unit instances and instances of subclasses of abcd:TypeSpecificInformation |
In the same way that DSW uses the object property
dsw:evidenceFor to link Tokens and Occurrences, ABCD uses the object property
abcd:hasTypeSpecificInformation to link
abcd:TypeSpecificInformation instances to Units. In addition, ABCD defines separate object properties that link an
abcd:Unit to instances of each subclass of
abcd:TypeSpecificInformation. To find all of those properties, I ran
this query; the specific object properties are all shown in the diagram above.
Clearly, the diagram above diagram is too complicated to insert as part of the man diagram comparing ABCD and DwC. Instead, I abbreviated it in the main diagram as shown in the following detail:
In this part of the diagram, I generalized the nine subclasses by a single bubble for the superclass abcd:TypeSpecificInformation. The link from the Unit to the evidence instance can be made through the abcd:hasTypeSpecificInformation or it can be made using one of the nine object properties that connect the Unit directly to the evidence.
In addition, I also placed abcd:MultimediaObject in the position of dsw:Token. Although images (and other kinds of multimedia) taken directly of the organism at the time the occurrence is recorded is often ignored by the museum community, with the flood of data coming from iNaturalist into GBIF, media is now a very important type of direct evidence for occurrences.
So in general, abcd:TypeSpecificInformation is synonymous with dsw:Token, with the exception that multimedia objects can serve as Tokens but aren't explicitly listed as subclasses of abcd:TypeSpecificInformation.
The place of controlled vocabularies in the model
The last major difference between the ABCD model and Darwin Core is how they deal with controlled vocabularies. Take for example the property abcd:hasSex. In the Wikibase installation, it's item Q1057 and has the range abcd:Sex. The range property would entail that abcd:Sex is a Class, but it's type is given in the Wikibase installation as Controlled Vocabulary rather than Class. As I mentioned earlier, in the abcd_concepts.owl ontology document, the controlled vocabularies are actually typed as owl:Class rather than skos:ConceptScheme as I would expect, with the controlled terms as instances of the controlled vocabularies.
So let's assume we have an abcd:Unit instance called _:occurrence1 that is a female. Using the model of ABCD, the following triples could describe the situation:
abcd:Sex a rdfs:Class.
abcd:hasSex a owl:ObjectProperty;
rdfs:range abcd:Sex.
abcd:Female a abcd:Sex;
rdfs:label "female"@en.
_:occurrence1 abcd:hasSex abcd:Female.
Currently, there are many terms in Darwin Core that say "Recommended best practice is to use a controlled vocabulary." However, most of these terms do not (yet) have controlled vocabularies, although this could change soon. Let's assume that the Standards Documentation Specification is followed and a SKOS-based controlled vocabulary identified by the IRI dwcv:gender is created to be used to provide values for the term dwciri:sex. Assume that the controlled vocabulary contains the terms dwcv:male and dwcv:female. The following triples could then describe the situation:
dwcv:gender a skos:ConceptScheme.
dwcv:female a skos:Concept;
skos:prefLabel "female"@en;
rdf:value "female";
skos:inScheme dwcv:gender.
_:occurrence1 dwc:sex "female".
_:occurrence1 dwciri:sex dwcv:female.
From the standpoint of generic modeling, neither of these approaches are "right" or "wrong". However, the latter approach is consistent with sections 4.1.2, 4.5, and 4.5.4 of the
TDWG Standards Documentation Specification as well as the pattern noted for controlled vocabularies in
section 8.9 of the W3C
Data on the Web Best Practices recommendation.
One reason that the ABCD graph diagram is more complicated than the DSW graph diagram is that some classes shown on the ABCD diagram as yellow bubbles (abcd:RecordBasis and abcd:Sex) and other classes not shown (like abcd:PermitType, abcd:NomenclaturalCode, etc.) represent controlled vocabularies rather than classes of linked resources.
Final thoughts
I have to say that I was somewhat surprised at how similar the ABCD and Darwin-SW graph models were. Perhaps I shouldn't be that surprised, given the DSW model's roots in the ACS model - it generally reflects the way the collections community views the universe and that view undoubtedly informs the ABCD model as well. That's good news, because it means that it should be possible to create a consensus graph model for Darwin Core and ABCD with minimal changes to either standard.
With such a model, it should be possible using SPARQL CONSTRUCT queries mediated by software to perform automated conversions from Darwin Core linked data to ABCD linked data. The CONSTRUCT query could insert blank nodes in places where the ABCD model has classes that aren't included in DwC. The conversion in the other direction would be more difficult since classes included in ABCD that aren't in DwC would have to be eliminated to make the conversion, and that might result in data loss as the data were denormalized. Still, the idea of any automated conversion is an encouraging thought!
The other thing that is clear to me from this investigation is that the current DwC and ABCD vocabularies could relatively easily be further developed into QL-like ontologies. That's basically what has already been done in the
abcd_concepts.owl ontology document and in DSW. It has been suggested that TDWG ontology development be carried out using the OBO Foundry system, but that system is designed to create and maintain EL-like ontologies. Transforming Darwin Core and ABCD to EL-like ontologies would be be much more difficult and it is not clear to me what would be gained by that, given that the primary use case for ontology development in TDWG would be to facilitate querying of large volumes of instance data.
For further reference
Ontologies vs. controlled vocabularies
The distinction between ontologies and controlled vocabularies is discussed in several standards:
To paraphrase these references, there is a fundamental difference between ontologies and controlled vocabularies.
Ontologies define knowledge related to some shared conceptualization in a formal way so that machines can carry out reasoning. They aren't primarily designed for human interaction.
Controlled vocabularies are designed to help humans use natural language to organize and find items by associating consistent labels with concepts. Controlled vocabularies don't assert axioms or facts. A
thesaurus (sensu
ISO 25964) is a subset of controlled vocabulary where its concepts are organized with explicit relationships (e.g. broader, narrower, etc.).
The
Data on the Web Best Practices recommendation notes in section 8.9 that controlled vocabularies and ontologies can be used together when the concepts defined in the controlled vocabulary are used as values for a property defined in an ontology. It gives the following example: "A concept from a thesaurus, say, 'architecture', will for example be used in the subject field for a book description (where 'subject' has been defined in an ontology for books)."
Kinds of ontologies
The
Introduction of the W3C OWL 2 Web Ontology Language Profiles Recommendation describes several profiles or sublanguages of the OWL 2 language for building ontologies. These profiles place restrictions on the structure of OWL 2 ontologies in ways that make them more efficient for dealing with data of different sorts. The nature of these restrictions are very technical and way beyond the scope of this post, but I mention the profiles because they provide a convenient way the characterize ontology modeling approaches. (I also refer you to
this post, which offers a very succinct description of the difference in the profiles.)
OWL 2 EL is suitable for "applications employing ontologies that define very large numbers of classes and/or properties". A classic example of such an ontology is the
Gene Ontology, where the data themselves are represented as tens of thousands of classes.
OWL 2 QL is suitable for "applications that use large volumes of instance data, and where query answering is the most important reasoning activity." A classic example of such an ontology is the
GeoNames ontology, which contains only 7 classes and 28 properties, but is used with over eleven million place feature instances. In OWL 2 QL, query answering can be implemented using conventional relational database systems.
I refer to ontologies with many classes and properties for which OWL 2 EL is suitable as "
EL-like ontologies", and ontologies with few classes and properties used with lots of instance data for which OWL 2 QL is suitable as "
QL-like ontologies".
Vocabularies and terms
Section 8.9 of the W3C Data on the Web Best Practices Recommendation describes vocabularies and terms in this way:
Vocabularies define the concepts and relationships (also referred to as “terms” or “attributes”) used to describe and represent an area of interest. They are used to classify the terms that can be used in a particular application, characterize possible relationships, and define possible constraints on using those terms. Several near-synonyms for 'vocabulary' have been coined, for example, ontology, controlled vocabulary, thesaurus, taxonomy, code list, semantic network.
So a vocabulary is a broad category that includes both ontologies and controlled vocabularies, and it is a collection of terms. In this post, I use "vocabulary" and "term" in this context and avoid using the word "
concept" unless I specifically mean it in the sense of a
skos:Concept (i.e. a term in controlled vocabulary).
Note: this was originally posted 2019-06-11 but was edited on 2019-06-12 to clarify the position of the subclasses of abcd:hasTypeSpecificInformation in the model.