Image by "Peng" from Wikimedia Commons CC BY-SA
When I was a kid, one of the most exciting things was to talk about and show my friends the new toys that I had just gotten. As an adult, it's no different - the toys are just different. Recently, I've been able to play with several new biodiversity informatics "toys" and I'm going to "show my friends" by writing several blog posts about them and let my friends (you) play with them if you want.
My first new toy: Darwin-SW version 0.4
Well, it's a little bit of an exaggeration to call the Darwin-SW ontology (DSW) a "new" toy. Cam Webb and I put the first version of it together in a few frenetic months back in 2010-2011. But DSW was in limbo for several years while we waited for some basic questions about the future of Darwin Core (DwC) to be settled by TDWG (Biodiversity Information Standards). One issue was what to do about the fact that DwC originally defined two parallel sets of classes, one in the main dwc:[1] namespace and another in the dwctype: [2] namespace. At first we thought the dwc: ones were the ones to use. But then we thought the dwctype: ones were preferred. Then we just weren't sure.
In the end (October 2014), the dwctype: classes were deprecated and the dwc: ones (including the new dwc:Organism class, which we had modeled in DSW) prevailed. So we finally knew what to build DSW around. The other thing that finally got settled in March of 2015 was the adoption of the DwC RDF Guide, which had languished in TDWG purgatory for a couple years. There were some properties that we had defined in DSW that would be unnecessary if they could be replaced by properties in the new dwciri: [3] namespace that the RDF Guide established. When the Guide was adopted, we were pretty much able to finalize the cast of characters in the DSW ontology.
I'm not going to say a lot about DSW here. There is more than most people would ever want to read about it at our old Google Code site, which we are now in the process of moving over to the new DSW GitHub site. The most succinct summary is in our manuscript submitted to the Semantic Web Journal, which has suffered in reviewer hell [4] for something like 15 months now since it was first submitted (shame on you SWJ!).
The bottom line here is that in the spring of this year we were finally able to release what we believe to be a stable version (0.4) of DSW and start to use it in a stable way in our own work. Which leads me to...
My second new toy: Bioimages RDF
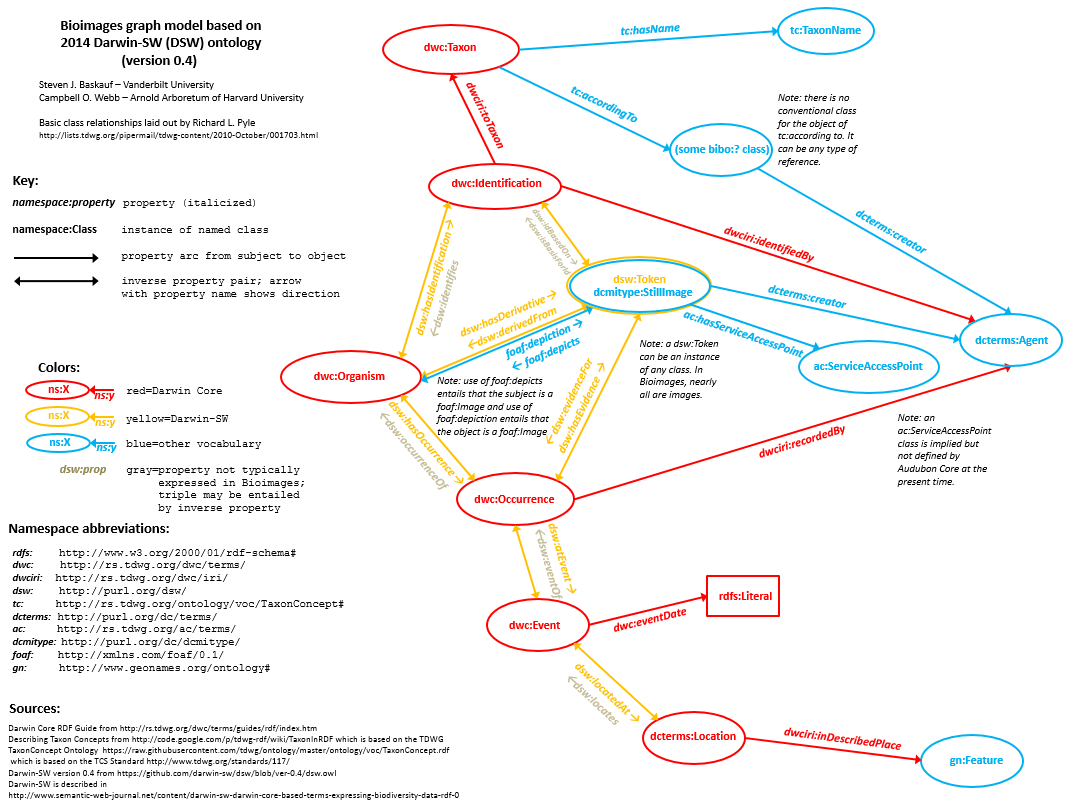
Again, it is a bit disingenuous to call Bioimages RDF "new". The Bioimages website had been serving some form of RDF/XML through content negotiation of its URIs since 2010. But the graph was unstable because the predicates were constantly changing as our understanding of how Darwin Core properties should be used as RDF and as DSW (on which the Bioimages graph model is primarily based) developed. When DSW 0.4 was at last released as a stable version, it was finally possible to release Bioimages RDF without potential users having to worry about the predicates and classes changing without warning.Click here to see a larger version of this graph
The Bioimages RDF graph model (diagram above) is basically the same as the DSW model, except that Bioimages adds properties from the Audubon Core Standard and the (somewhat obsolete but not yet replaced) TDWG TaxonConcept Ontology.
The actual Bioimages dataset itself is not as complicated as the RDF graph model shown above. The data actually "live" primarily in five pipe-delineated CSV files, one each for agents, determinations, images, organisms, and sensu references. The data are CC0 licensed and freely available on the Bioimages GitHub site. The five resource types that Bioimages documents in the CSV files would only require five of the nodes (bubbles) in the diagram above. So why make it more complicated?
The point of Darwin-SW is to create the simplest graph model that enables all of the one-to-many relationships that most people in the traditional biodiversity informatics community have said they need. (I said "traditional" because the DSW model doesn't do much for metagenomics people. But that wasn't the problem we set out to address.) In the simplest possible model, Bioimages doesn't really "need" Event and Location nodes because all Occurrences are considered to be at separate Events and Locations (i.e. there is a 1:1:1 relationship between Occurrence, Event, and Location). But when the data in the CSV files are expressed as RDF, the "extra" nodes are created so that it is possible for the Bioimages data to be merged with data from a provider that has 1:many relationships in places where Bioimages doesn't (for example a provider that's tracking many Occurrences at one collecting Event, or many Events at one Location). Just for fun, Cam once merged his Xmalesia data from Indonesia (expressed as DSW RDF) with the Bioimages data (mostly from the southeastern US). He had no problem running simple queries on the combined dataset because both followed the basic DSW graph model.
The 1488578 RDF triples currently in the Bioimages dataset are generated from the CSV files. In the past I did it using some horrid software that I wrote myself. However, recently I've started hanging out with a fun group of people who are mostly affiliated with the Jean and Alexander Heard Library at Vanderbilt. Their interest is primarily in the Digital Humanities (I had to look that up - see my blog post on the subject), but we have a common interest in playing with metadata. The primary outcome of this interaction for me was that I have started to learn how to use XQuery, so as an exercise I wrote queries that would transform the data in the CSV files into XHTML (for the Bioimages website; view source for this file for an example) and RDF/XML (to generate the RDF triples in the Bioimages graph; here is an example) files for organisms and images. Currently, the queries output into many static files that are served in a boring way when the URIs are dereferenced via content negotiation, but along with the XQuery group I've also been experimenting with the server capabilities of BaseX, so eventually these individual files may be generated on the fly from a database. Using a hack of those XQueries, I can also output the entire database as RDF/XML into a few files that zip up into a relatively small and portable file (bioimages-rdf.zip) that can be obtained from the Bioimages GitHub repo. The version in the current database release (2014-07-22; http://dx.doi.org/10.5281/zenodo.21009) takes up only 3.7 MB.
As fun as it is to say that I've generated an RDF graph of about 1.5 million triples representing real data, those triples are completely useless unless you can do something with them. So that brings me to ...
My third new toy: Callimachus-based SPARQL endpoint
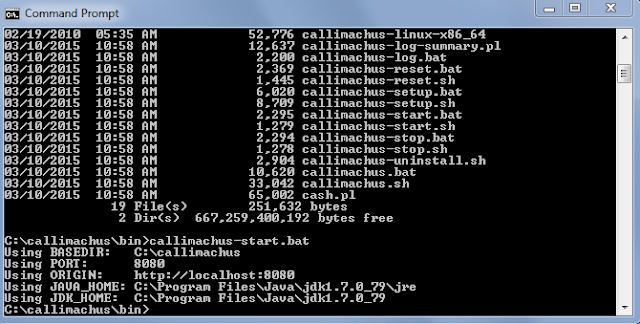
Right around the time last winter when things were starting to move with the stalled TDWG stuff, Suellen Stringer-Hye , Digital Librarian at the Heard Library approached me about proposing a Dean's Fellow project to explore using Callimachus to set up a SPARQL endpoint that could query a triplestore containing the Bioimages RDF. I was very excited about this prospect because I was keen to find out what it was possible to do with a real RDF dataset based on Darwin-SW. The project was approved and Sean King, a senior in the Vanderbilt Biological Sciences Department who works on genomics in the Rokas Lab, was chosen to work on the the project with us as a Dean's Fellow.Callimachus is actually really easy to set up as a localhost on a desktop machine. The installation directions on the Callimachus website are very straightforward. I've set it up on two of my Windows computers and it only took a few minutes to get it running, although it may take significantly longer if you run into port conflicts and have to troubleshoot that. The server is cranked up by running a batch file from the DOS command prompt and you are up and running:
You run the software through a browser pointed to the localhost (default: port 8080). That takes you to a screen where you can upload data files or invoke the SPARQL endpoint:
Before you can start playing with the SPARQL endpoint, you have to upload the RDF data files. Callimachus will accept RDF in a number of serializations. I used XML because that's what I had, but it will also accept RDF in Turtle and JSON-LD (I think) serializations. It takes only a few moments to upload a small file, such as the FOAF vocabulary. However, it can take several minutes to upload larger files, such as the Bioimages images.rdf file that contains hundreds of thousands of triples. On my first try loading that file, I thought that I'd hung the computer but all that was required was patience. It is also possible to load an entire batch of XML files that are bundled in a zip file. I haven't tried this, but Suellen says it works fine. So one should be able to load the entire Bioimages dataset just by uploading the bioimages-rdf.zip file into Callimachus. If you upload a new version of a particular file, Callimachus will replace any previous version of the file that has the same name.
The names of the files that contain particular triples are significant because Callimachus uses them when referencing particular dataset graphs in SPARQL queries. For example, if a query contains the line:
FROM <http://localhost:8080/organisms.rdf>
the triples contained in the organisms.rdf file will be searched in the query. If you have only loaded a single dataset into your Callimachus instance, it doesn't really matter if your query restricts the search via FROM clauses. However, if several datasets are loaded, queries may return annoying results from datasets that are irrelevant to the search if you don't specify FROM clauses.
Once you've loaded all of the files containing the triples, just click on the SPARQL link in the file list and voilà! ... you are staring at a blank box that expects you to enter a SPARQL query. If you are unfamiliar with SPARQL, this is a bit daunting. In my next blog post, I'm going to provide some queries that you can run on the Bioimages data.
After playing around with Callimachus as a localhost for a while, Sean, Suellen, and I started talking about the next step, which was to get Callimachus running on the library server. When you have a new toy, it is always nice to let others play with it, so we wanted to make sure that it was possible for queries to be sent from clients outside the rdf.library.vanderbilt.edu subdomain. Otherwise, users would be stuck with typing ugly queries into the blank box. The issue is to change the settings on Callimachus to allow cross-domain requests. It took us a while to find where the settings were located, but eventually we figured it out:
1. In the Callimachus Home folder, click Edit and add "*" as an Allowed origin.
2. Load the SPARQL page (http://localhost:8080/sparql?view), drop down the menu, select "Permissions", and change the "Reader" value to "public".
If you want to experiment with querying a Callimachus endpoint via a script in a webpage, it is necessary to make these changes even if you aren't planning to put the endpoint on the Internet. By default, most browsers are using port 80, so if a browser queries the server (at port 8080), it's considered a cross-domain request and the endpoint will refuse to respond if the Callimachus settings are left at their defaults. (I hope I have stated the technical details of this correctly. Many thanks to Cliff Anderson, Heard Library Director of Scholarly Communications, for helping us troubleshoot this problem!)
Playing (nicely) with the toys
We have just recently finished creating a somewhat user-friendly demo web page that queries the Bioimages RDF graph via the Heard Library public SPARQL endpoint. You can try it at:http://www.library.vanderbilt.edu/webimages/Bioimages/
(Note: we are still tinkering with this page, so its behavior may be somewhat unpredictable. Many thanks to Jodie Gambill, Systems Librarian at the Heard Library for helping us get it to work the way we wanted!) To the end user, the page looks like a typical search page that uses web services to provide appropriate options to the user. By selecting appropriate values from the dropdowns, a user can screen images and view thumbnails of images that meet the selected criteria. Clicking on a thumbnail takes the user to the image web page where detailed metadata and access to the original high-resolution image are provided.
Under the hood, this behavior is accomplished in an atypical way: by making queries via Javascript in the HTML to the public SPARQL endpoint, and using the results to change the appearance of the web page. Because the page is being served from http://www.library.vanderbilt.edu/ and the endpoint is at http://rdf.library.vanderbilt.edu/, the queries are being made cross-domain. Because we have allowed cross-domain queries, the HTML page could actually be served from anywhere (even loaded as a local file on your hard drive) and it would still work.
The "guts" of the page are in the Javascript file http://www.library.vanderbilt.edu/webimages/Bioimages/bioimages.js, which must be located in the same directory as the HTML. The basic mechanism that makes the page work involves several steps:
1. Assemble a string that contains the query and URL-encode it. For example:
var string = "PREFIX Iptc4xmpExt: <http://iptc.org/std/Iptc4xmpExt/2008-02-29/>"+
"PREFIX rdfs: <http://www.w3.org/2000/01/rdf-schema#>"+
'SELECT DISTINCT ?category WHERE {' +
"?image Iptc4xmpExt:CVterm ?view." +
"?view rdfs:subClassOf ?featureCategory." +
"?featureCategory rdfs:label ?category." +
'}'
+'ORDER BY ASC(?category)';
var encodedQuery = encodeURIComponent(string);
2. Append the string to the URI of the endpoint as a query, then use AJAX (facilitated by jQuery) to send the query to the endpoint and receive the results XML:
$.ajax({
type: 'GET',
url: 'http://rdf.library.vanderbilt.edu/sparql?query=' + encodedQuery,
headers: {
Accept: 'application/sparql-results+xml'
},
success: parseCategoryXml
I really didn't want to get the results in XML format. There is a JSON format for SPARQL results that can be requested in the Accept header:
application/sparql-results+json
but unfortunately XML is the only format currently supported by Callimachus. To see an example of the result of a query, click here.
3. Use jQuery's XML-processing capabilities to pull the desired values from the results XML and insert them into a component of the page's HTML:
$("#div1").html("").append(resultsStatement + "<table>");
//step through each "result" element
$(xml).find("result").each(function() {
tableRow="<tr><td>";
// pull the "binding" element that has the name attribute of "image"
$(this).find("binding[name='image']").each(function() {
tableRow=tableRow+"<a href='"+$(this).find("uri").text() + "'>";
});
// pull the "binding" element that has the name attribute of "uri"
$(this).find("binding[name='uri']").each(function() {
tableRow=tableRow+"<img src='"+$(this).find("uri").text() + "'></a></td><td>";
});
// pull the "binding" element that has the name attribute of "title"
$(this).find("binding[name='title']").each(function() {
tableRow=tableRow+$(this).find("literal").text() + "</td></tr>";
$("#div1").append(tableRow);
});
});
$("#div1").append("</table>");
We hacked these basic three functions several times to make the page behave the way we wanted. First, after the page loads, the endpoint is queried to find out what values of genus, state, and category exist in the database. Those values are used to populate the corresponding three dropdowns. If the user selects a value for genus, the endpoint is further queried to find out what species values exist in the database for that genus and the values of the species dropdown are set. When the "Search" button is clicked, the main query is sent and the results are formatted into an HTML table that is displayed at the bottom of the page.
This is a relatively simple application, but it demonstrates how anyone anywhere could create code to retrieve data from the Heard Library public SPARQL endpoint and use it to do something interesting in a Web page. Even more interesting things could be done with more user interaction to create more complex queries or by displaying the results in other ways, such as maps or charts.
Next time: playing roughly with the toys
The demo webpage is a nice demonstration of making use of the three toys (Darwin-SW, the Bioimages data, and the Heard Library SPARQL endpoint) to do something useful. However, whenever kids get a new toy, they are usually not interested in playing with it according to the instructions. In my next blog post, I will report on my attempts to try doing more interesting things with the toys, including finding out what it takes to break them.[1] dwc: = http://rs.tdwg.org/dwc/terms/
[2] dwctype: = http://rs.tdwg.org/dwc/type/
[3] dwciri: = http://rs.tdwg.org/dwc/iri/
[4] Possibly the only thing worse than TDWG purgatory!


No comments:
Post a Comment